Abstract
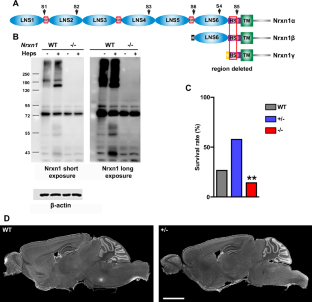
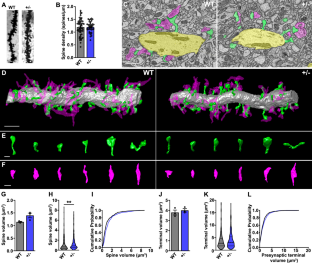
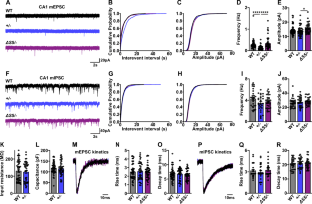
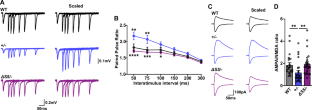
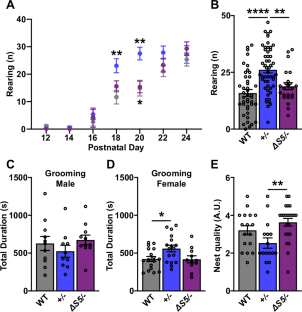
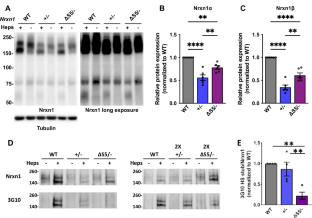
NRXN1 encoding the synaptic organizing protein neurexin 1 (Nrxn1) is among the strongest risk genes for autism spectrum disorders as well as other neuropsychiatric disorders. The most common contributing mutation is a deletion in one allele. While mice lacking one form of the protein, Nrxn1α, have been characterized, information is lacking on animal models with heterozygous deletion of all isoforms, as well as on therapeutic approaches directly targeting Nrxn1. We report that Nrxn1+/− mice with a deletion affecting all isoforms, α, β and γ, show deficits in excitatory synaptic transmission affecting presynaptic and postsynaptic properties at hippocampal CA3-CA1 synapses, and show increased repetitive behaviors. Based on previous studies indicating that exclusion of the insert at Nrxn1 splice site 5 (S5) boosts synaptic transmission, we tested S5 exclusion as a therapeutic approach. Genetic exclusion of S5 in the remaining Nrxn1 allele alleviated the deficits, restoring miniature excitatory postsynaptic current frequency, paired pulse ratio, AMPA/NMDA ratio, and repetitive behaviors to wild type levels and partially restoring Nrxn1 protein level in Nrxn1ΔS5/- compared to Nrxn1+/− mice. These data suggest that S5 exclusion may be a beneficial therapeutic direction in cases of neuropsychiatric disorders involving NRXN1.
This is a preview of subscription content, access via your institution
Access options
Access to this article via ICE Institution of Civil Engineers is not available.
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Data availability
All data relevant to evaluate the findings of this study are available in the main paper or supplementary material.
References
Sudhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–69.
Reissner C, Runkel F, Missler M. Neurexins. Genome Biol. 2013;14:213.
Gomez AM, Traunmuller L, Scheiffele P. Neurexins: molecular codes for shaping neuronal synapses. Nat Rev Neurosci. 2021;22:137–51.
Bena F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of nrxn1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403.
Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, et al. Investigation of nrxn1 deletions: clinical and molecular characterization. Am J Med Genet A. 2013;161A:717–31.
Castronovo P, Baccarin M, Ricciardello A, Picinelli C, Tomaiuolo P, Cucinotta F, et al. Phenotypic spectrum of nrxn1 mono- and bi-allelic deficiency: a systematic review. Clin Genet. 2020;97:125–37.
Hu Z, Xiao X, Zhang Z, Li M. Genetic insights and neurobiological implications from nrxn1 in neuropsychiatric disorders. Mol Psychiatry. 2019;24:1400–14.
Cosemans N, Vandenhove L, Vogels A, Devriendt K, Van Esch H, Van Buggenhout G, et al. The clinical relevance of intragenic nrxn1 deletions. J Med Genet. 2020;57:347–55.
Flaherty E, Zhu S, Barretto N, Cheng E, Deans PJM, Fernando MB, et al. Neuronal impact of patient-specific aberrant nrxn1alpha splicing. Nat Genet. 2019;51:1679–90.
Fuccillo MV, Pak C. Copy number variants in neurexin genes: phenotypes and mechanisms. Curr Opin Genet Dev. 2021;68:64–70.
Sebastian R, Jin K, Pavon N, Bansal R, Potter A, Song Y, et al. Schizophrenia-associated nrxn1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat Commun. 2023;14:3770.
Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in nrxn1. Cell Stem Cell. 2015;17:316–28.
Pak C, Danko T, Mirabella VR, Wang J, Liu Y, Vangipuram M, et al. Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived nrxn1-mutant neurons. Proc Natl Acad Sci USA. 2021;118:e2025598118.
Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–8003.
Xu B, Ho Y, Fasolino M, Medina J, O’Brien WT, Lamonica JM, et al. Allelic contribution of nrxn1alpha to autism-relevant behavioral phenotypes in mice. PLoS Genet. 2023;19:e1010659.
Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS ONE. 2013;8:e67114.
Dachtler J, Ivorra JL, Rowland TE, Lever C, Rodgers RJ, Clapcote SJ. Heterozygous deletion of alpha-neurexin i or alpha-neurexin ii results in behaviors relevant to autism and schizophrenia. Behav Neurosci. 2015;129:765–76.
Dai J, Aoto J, Sudhof TC. Alternative splicing of presynaptic neurexins differentially controls postsynaptic nmda and ampa receptor responses. Neuron. 2019;102:993–1008.
Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic ampa receptor trafficking. Cell. 2013;154:75–88.
Lu H, Zuo L, Roddick KM, Zhang P, Oku S, Garden J, et al. Alternative splicing and heparan sulfation converge on neurexin-1 to control glutamatergic transmission and autism-related behaviors. Cell Rep. 2023;42:112714.
Zhang P, Lu H, Piexoto RT, Pines MK, Ge Y, Oku S, et al. Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell. 2018;174:1450–64.
Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, et al. Trakem2 software for neural circuit reconstruction. PLoS ONE. 2012;7:e38011.
Blaney CE, Gunn RK, Stover KR, Brown RE. Maternal genotype influences behavioral development of 3xtg-ad mouse pups. Behav Brain Res. 2013;252:40–8.
Dierssen M, Fotaki V, Martinez de Lagran M, Gratacos M, Arbones M, Fillat C, et al. Neurobehavioral development of two mouse lines commonly used in transgenic studies. Pharmacol Biochem Behav. 2002;73:19–25.
Fox WM. Reflex-ontogeny and behavioural development of the mouse. Anim Behav. 1965;13:234–41.
Tremml P, Lipp HP, Muller U, Ricceri L, Wolfer DP. Neurobehavioral development, adult openfield exploration and swimming navigation learning in mice with a modified beta-amyloid precursor protein gene. Behav Brain Res. 1998;95:65–76.
Hill JM, Lim MA, Stone MM Developmental milestones in the newborn mouse. in: Gozes I, Editor. Neuropeptide techniques. Humana Press; Totowa, New Jersey, 2008. p. 131–49.
Friard OGamba M. BORIS: a free, versatile open-source event-logging software for video/audio coding and live observations. Methods Ecol Evol. 2016;7:1325–30.
Kalueff AV, Aldridge JW, LaPorte JL, Murphy DL, Tuohimaa P. Analyzing grooming microstructure in neurobehavioral experiments. Nat Protoc. 2007;2:2538–44.
Deacon RM. Assessing nest building in mice. Nat Protoc. 2006;1:1117–9.
Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507.
Uchigashima M, Cheung A, Suh J, Watanabe M, Futai K. Differential expression of neurexin genes in the mouse brain. J Comp Neurol. 2019;527:1940–65.
Trotter JH, Hao J, Maxeiner S, Tsetsenis T, Liu Z, Zhuang X, et al. Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. J Cell Biol. 2019;218:2677–98.
Harris KM, Kater SB. Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci. 1994;17:341–71.
Gandhi T, Lee CC. Neural mechanisms underlying repetitive behaviors in rodent models of autism spectrum disorders. Front Cell Neurosci. 2020;14:592710.
Kim H, Lim CS, Kaang BK. Neuronal mechanisms and circuits underlying repetitive behaviors in mouse models of autism spectrum disorder. Behav Brain Funct. 2016;12:3.
Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, et al. Alpha-neurexins couple ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–48.
Keck T, Keller GB, Jacobsen RI, Eysel UT, Bonhoeffer T, Hubener M. Synaptic scaling and homeostatic plasticity in the mouse visual cortex in vivo. Neuron. 2013;80:327–34.
Hobbiss AF, Ramiro-Cortes Y, Israely I. Homeostatic plasticity scales dendritic spine volumes and changes the threshold and specificity of hebbian plasticity. iScience. 2018;8:161–74.
Luo F, Sclip A, Jiang M, Sudhof TC. Neurexins cluster ca(2+) channels within the presynaptic active zone. EMBO J. 2020;39:e103208.
Budreck EC, Kwon OB, Jung JH, Baudouin S, Thommen A, Kim HS, et al. Neuroligin-1 controls synaptic abundance of nmda-type glutamate receptors through extracellular coupling. Proc Natl Acad Sci USA. 2013;110:725–30.
Bhouri M, Morishita W, Temkin P, Goswami D, Kawabe H, Brose N, et al. Deletion of LRRTM1 and LRRTM2 in adult mice impairs basal ampa receptor transmission and ltp in hippocampal ca1 pyramidal neurons. Proc Natl Acad Sci USA. 2018;115:E5382–9.
Dhume SH, Connor SA, Mills F, Tari PK, Au-Yeung SHM, Karimi B, et al. Distinct but overlapping roles of LRRTM1 and LRRTM2 in developing and mature hippocampal circuits. eLife. 2022;11:e64742.
Dai J, Patzke C, Liakath-Ali K, Seigneur E, Sudhof TC. GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature. 2021;595:261–5.
Dai J, Liakath-Ali K, Golf SR, Sudhof TC. Distinct neurexin-cerebellin complexes control ampa- and nmda-receptor responses in a circuit-dependent manner. eLife. 2022;11:e78649.
Nozawa K, Sogabe T, Hayashi A, Motohashi J, Miura E, Arai I, et al. In vivo nanoscopic landscape of neurexin ligands underlying anterograde synapse specification. Neuron. 2022;110:3168–85.
Matsuda K, Budisantoso T, Mitakidis N, Sugaya Y, Miura E, Kakegawa W, et al. Transsynaptic modulation of kainate receptor functions by c1q-like proteins. Neuron. 2016;90:752–67.
Hauser D, Behr K, Konno K, Schreiner D, Schmidt A, Watanabe M, et al. Targeted proteoform mapping uncovers specific neurexin-3 variants required for dendritic inhibition. Neuron. 2022;110:2094–109.
Mercuri E, Pera MC, Scoto M, Finkel R, Muntoni F. Spinal muscular atrophy - insights and challenges in the treatment era. Nat Rev Neurol. 2020;16:706–15.
Wirth B. Spinal muscular atrophy: in the challenge lies a solution. Trends Neurosci. 2021;44:306–22.
Furlanis E, Traunmuller L, Fucile G, Scheiffele P. Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat Neurosci. 2019;22:1709–17.
Lukacsovich D, Winterer J, Que L, Luo W, Lukacsovich T, Foldy C. Single-cell rna-seq reveals developmental origins and ontogenetic stability of neurexin alternative splicing profiles. Cell Rep. 2019;27:3752–9.
Boxer EE, Seng C, Lukacsovich D, Kim J, Schwartz S, Kennedy MJ, et al. Neurexin-3 defines synapse- and sex-dependent diversity of gabaergic inhibition in ventral subiculum. Cell Rep. 2021;37:110098.
Acknowledgements
We thank Xiling Zhou for excellent technical assistance, and Sam Barnes, Michael Bellefontaine, Spencer Brown, Val Burdeyny, Emre Fertan, Jessica Garden, Hiyam Gedalia, Elias Habib, Hugh McClelland, Ella Puolitaipale, Qendresa Sahiti, Oliver Schnare, Katie Scott, Aimée Wong, and Jessica Zhang for assistance with behavioural testing. We thank Ed Parker (Vision Core) and Sharmon Knecht, both of the University of Washington, for technical assistance with electron microscopy. Funding was provided by Simons Foundation Autism Research Initiative SFARI 608066 to A.M.C. and R.E.B., Canadian Institutes of Health Research PJT-183943 and FDN-143206 to A.M.C., Natural Sciences and Engineering Research Council of Canada A7441 to R.E.B., National Institutes of Health EYP30-01730 to M. Neitz, a Vanier Canada Graduate Scholarship to H.L., and a Brain Canada’s Shireen and Edna Marcus Excellence Award to H.L.
Author information
Authors and Affiliations
Contributions
The overall project was designed by HL, REB and AMC Experiments were designed, performed, and data analyzed by HL, YG and LZ under the oversight of AMC, by PZ for the knockout validation, by OL and KM under the oversight of HL and ROLW for the SBFSEM, and by KMR under the oversight of REB for the behavioral analyses. The manuscript was written by HL and AMC with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal procedures were approved by the Animal Care Committee at the University of British Columbia, protocols A16-0086, A20-0095, and A24-0094, and by the University Committee on Laboratory Animals at Dalhousie University, protocols 17–58, 19–072, 19–090, 21–043, 21–079, and 22–063. No human research was performed.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, H., Roddick, K.M., Ge, Y. et al. Targeted splicing approach for alleviation of a neurexin 1 haploinsufficiency model. Mol Psychiatry (2025). https://doi.org/10.1038/s41380-025-03017-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-025-03017-w



